|
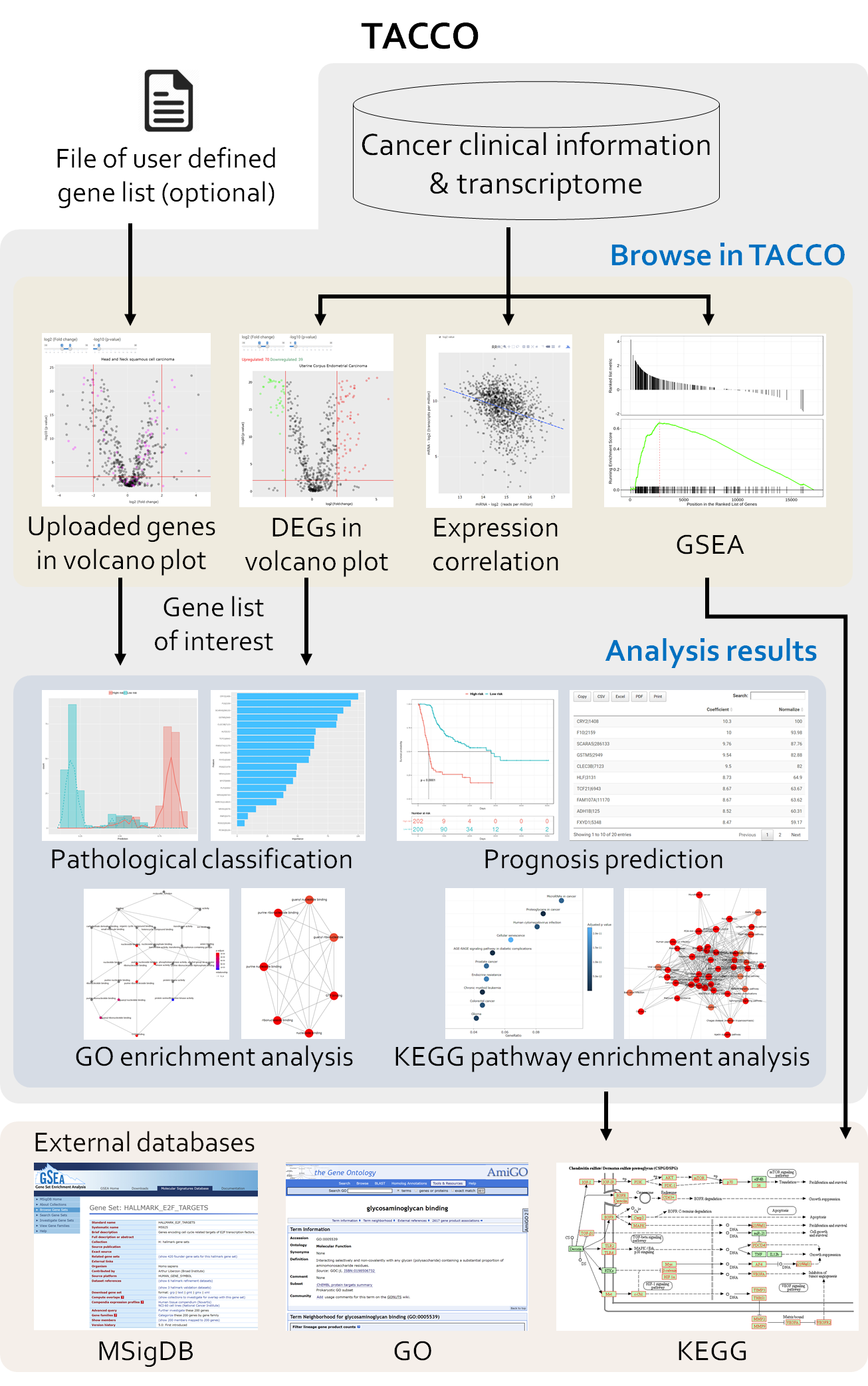
TACCO is a database designed specifically for connecting transcriptome alterations, pathway alterations and clinical outcomes in cancers. Users can either select differentially expressed genes with different criteria within the TACCO database or upload their own gene list to be analyzed. Users can then perform KEGG pathway enrichment analysis, Gene ontology analysis or construct a multiple gene model for the prognosis prediction using this gene list.
TACCO provides the followings:
1.) Browse the expression level of a specific gene/miRNA across different cancer types.
2.) Explore the correlations between the expression between miRNAs and their target genes.
3.) Search Gene Set Enrichment Analysis results to investigate transcriptome alterations in different cancer types.
4.) Identify differentially expressed genes (DEGs).
5.) Identify differentially expressed miRNAs (DEmiRNAs).
6.) Perform KEGG pathway enrichment analysis for a gene list of interest.
7.) Carry out GO enrichment analysis for a gene list of interest.
8.) Construct a prognosis prediction model for a gene list of interest in different cancer types.
9.) Construct a pathological classification model from a gene list of interest in different cancer types.
Multiple cancer types were included in TACCO: Bladder Urothelial Carcinoma (BLCA), Breast invasive carcinoma (BRCA), Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), Cholangiocarcinoma (CHOL), Esophageal carcinoma (ESCA), Head and Neck squamous cell carcinoma (HNSC), Kidney Chromophobe (KICH), Pan-kidney cohort (KICH+KIRC+KIRP) (KIPAN), Kidney renal clear cell carcinoma (KIRC), Kidney renal papillary cell carcinoma (KIRP), Liver hepatocellular carcinoma (LIHC), Lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC), Pancreatic adenocarcinoma (PAAD), Pheochromocytoma and Paraganglioma (PCPG), Prostate adenocarcinoma (PRAD), Skin Cutaneous Melanoma (SKCM), Stomach adenocarcinoma (STAD), Thyroid carcinoma (THCA), Thymoma (THYM), Uterine Corpus Endometrial Carcinoma (UCEC).
New users are advised to read our
tutorial
before using TACCO.
|














